Laurence Black, PhD
Postdoctoral Fellow
University of Alabama at Birmingham
Precision medicine is an exciting approach to treat diseases on an individual basis that considers and incorporates the genetic make-up, environment, and lifestyle of each patient to develop personalized therapeutic approaches. New, powerful technologies are emerging that allow for data collection to elucidate molecular and cellular mechanisms of pathology that could make this precise approach to patient care possible. This approach has been implemented in cancer biology, for example, whereby specific transcriptomic signature and tumor staining is used to determine treatment options. In nephrology, many diagnoses of renal pathologies rely on analyses of histological staining in biopsied tissue. More recently, state-of-the-art tissue interrogation such as three-dimensional imaging and tissue cytometry coupled with Volumetric Tissue Exploration and Analysis (VTEA), first described in the literature in 2017, has gained traction. Tarek Ashkar, MD at the Indiana Center for Biological Microscopy has optimized this technique that is termed “3DTC.” Dr. Ashkar’s group is able to utilize this technique to better understand the kidney and the relationships between the multitude of cells within this complex organ.
There has been an explosion of sophisticated transcriptomic analyses in nephrology research that allow for insights into changes in gene signature in disease at single-cell or single-nucleus resolution. While these techniques have uncovered precise mechanisms of kidney injury, suggested novel cell-cell interactions, and even discovered new cell types and states, valuable details such as cellular organization and structure, as well as physical interaction between cell types are often overlooked. Standard microscopic techniques only investigate a two-dimensional plane and are extremely limiting in evaluating spatial location and organization of structures within complex organ systems, such as the kidney. This is especially true when studying the specific dynamics of inflammatory infiltrate, vascular rarefaction, and fibrosis, among many other hallmarks of acute and chronic kidney disease. Just think of all of the information that can be gained by a technique that offers the ability to image an entire nephron, its associated vasculature, and cell-to-cell interaction, such as immune cells interacting with damaged structures!
Using confocal microscopy, a technique that allows for the manipulation of field depth and background in the focal plane, users are able to image serial optical sections and collect three-dimensional data, which can beautifully capture the cellular network of organs at subcellular resolution. In contrast, using flow cytometry or transcriptomics, researchers can detect the presence or absence of different types of immune cells. With 3DTC, researchers are able to detect different cell types in thorough detail, such as the specific spatial location of kidney resident macrophages. Depending on the mode of renal injury, the dynamics and spatial distribution of particular cells reveals a wealth of information concerning pathogenesis of disease. 3DTC can enhance investigators’ understanding of the pathogenesis of injury in these contexts by gaining otherwise lost information. For example, instead of interrogating the presence or absence of a particular cell type of interest, investigators can ask questions based on spatial location of a particular cell of interest or evaluate potential cell-cell interactions.
In order to process tissues for 3DTC (Workflow shown in Figure 1), samples must be frozen in optimal cutting temperature (OCT) compound, cut into 50 μm thick sections, and temporarily mounted. Using two-photon microscopy of label-free tissue, endogenous autofluorescence and signals from second harmonic generation imaging (SHG) can be detected. Autofluorescence is especially important to account for when imaging severely damaged tissue, especially in the kidney as autofluorescence is common in this organ. It is important to also note that autofluorescence can occur from NADH, collagen, elastin, and flavins. Importantly, SHG detects signals predominantly caused by fibrillary collagen.
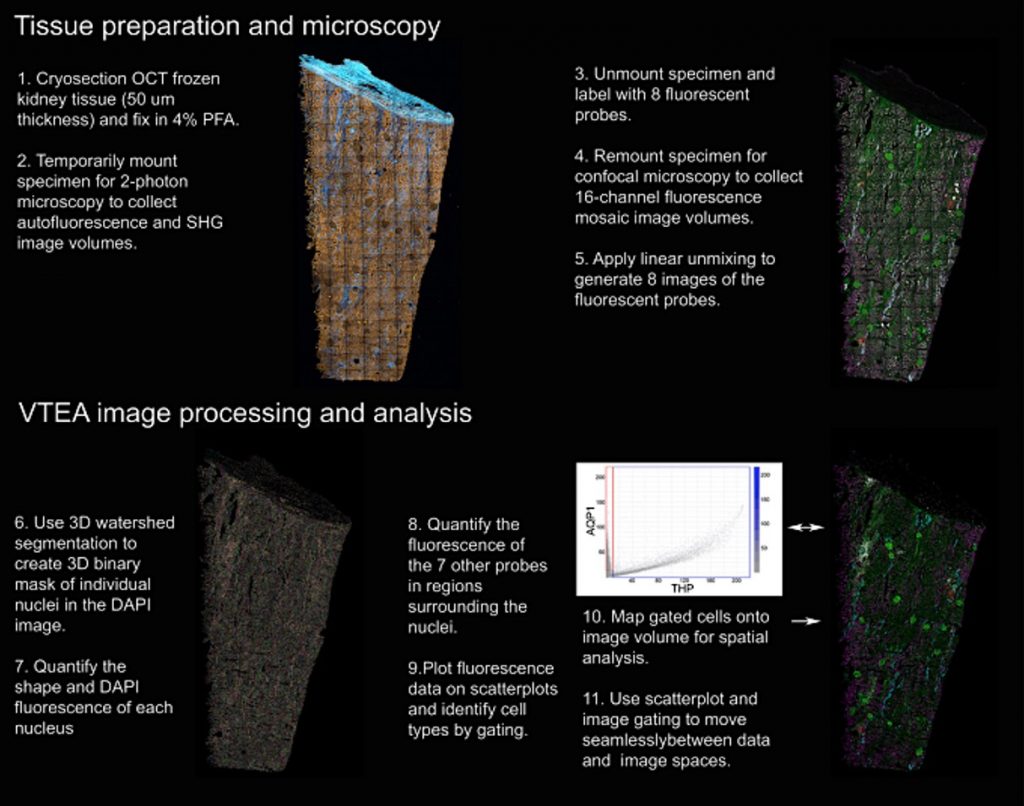
Tissue is then unmounted and further processed for immunofluorescent labeling with up to 8 antibodies (such as AQP1, CD68, THP—whatever you’d like!), including phalloidin (to label filamentous actin) and DAPI (for nuclei labeling). The sections are imaged to collect 16-channel fluorescence mosaic image volumes. These mosaics are constructed by smaller (20X) 400 μm image volumes. Images are then analyzed to pinpoint each of the 8 fluorescent markers, after which volumetric tissue exploration and analysis (VTEA) software (ImageJ plugin!) is used to segment the DAPI volume into individual nuclei. Fluorescent signal in each channel is converted into scatterplots for cell type gating, which can be mapped back to the original image. These scatter plots and image maps are used to explore the image for analysis.
Images are converted into quantitative data to most accurately analyze the patterns and distribution of cells, which is referred to as “tissue cytometry.” This is similar to flow cytometry, in that this method detects and quantifies fluorescent signals from individual cells; however, tissue cytometry includes spatial location data as well, allowing investigators to gain additional insight about cellular processes throughout the experiment. What is even more exciting about 3DTC is that the signal can be parsed out by VTEA analysis to account for fluorescent contamination, should the signal bleed into inappropriate channels.
Application of this technology can convey massive amounts of information that was previously lost using standard microscopy techniques. For example, Ferkowicz and colleagues were able to detect advanced fibrosis, neutrophil, macrophage, and T cell infiltrates, and even the cell density of glomeruli in human clinical biopsy samples from patients with diabetic nephropathy to demonstrate the power of 3DTC that could be exploited for personalized medicine. Furthermore, Winfree and colleagues highlighted the use of 3DTC for understanding the distribution of immune cells in both human and mouse kidney tissue, highlighting the power of this approach for understanding the pathogenesis of disease.
3DTC is emerging as a sophisticated approach to understand complex cellular dynamics, and continues to be refined for ease of application. Its use complements other molecular techniques, such as flow cytometry and transcriptomics in investigating scientific questions, but stands alone as a powerful tool to understand complex biological dynamics in kidney health and disease.
Reviewed by: Elinor Mannon, Tanecia Mitchell, PhD, Matthew Sparks, MD, FASN, FAHA


